The IDB SNP chip has been used for nearly all of Ireland’s cattle genotyping and was developed by ICBF and its research collaborators, Teagasc etc., for our farmers to lower the cost of parentage verification and contribute to genetic breeding values. Along the way we’ve also been adding a number of SNPs* that test for disease causing (or preventing) alleles, there are also a number of genes that contribute to milk or meat phenotypes in animals and finally we have a number of “discovery” SNPs. These are ones where Irish farmers will play an important role in helping us determine how useful these are in a large cattle population. Dr. Matthew McClure collaborated with a number of researchers across the globe when developing the chip and putting these discovery SNPs on the chip. The idea is if a researcher finds a SNP of interest that they think may contribute to a cows susceptibility to a disease like mastitis, or displaced-abomasum but are not sure if it is found in many breeds, just their research population, or possibly just a one-off fluke in the research, we can put it on the SNP chip and analyse it via health records that farmers have given ICBF access to. The more records the better! Unfortunately we’ve seen a drop in the number of farms recording health and disease data to ICBF in the past 2 years, so our work towards this has slowed considerably. It takes a large number of animals to be sure these SNPs are useful. When we have determined if these SNPs are useful indicators of susceptibility to disease, we can report them back to farmers.
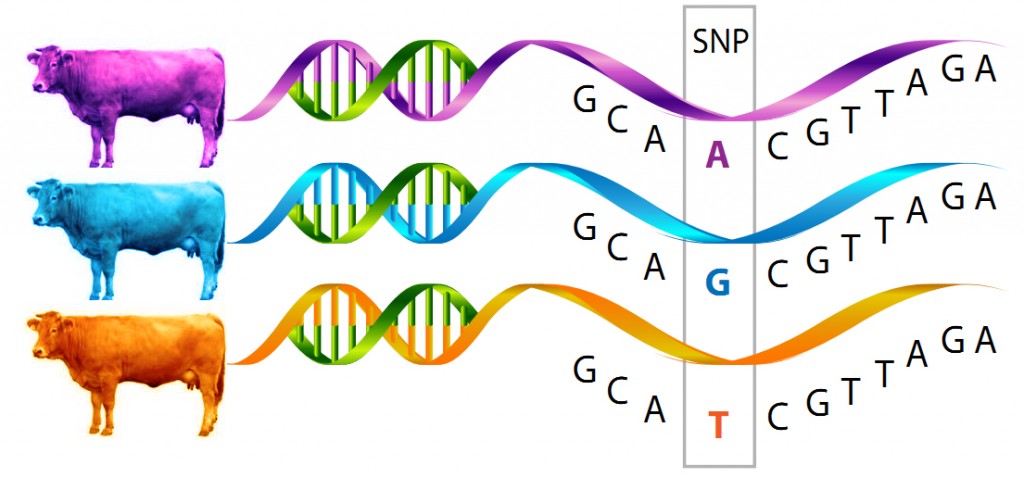
Right now we are working on a way to report to farmers what gene variants their animals are carrying so that when choosing a bull, they can determine not only what positive traits the bull has, but also evaluate the possibility of a bull passing on a known genetic defect or other genes to their offspring. The best part about this approach is that even though an animal might be a carrier for a disease, we don’t have to rule the animal out as a parent of the next generation because it may have many other good traits, we just have to be more selective on who we choose to mate with that animal. If you mate a carrier and an animal without the “bad” gene variant you will not get an affected offspring. It’s all about being selective with your mating. This will also allow farmers to introduce specific good genes into their herd where they may not have had them before.
This work goes hand in hand with the Genetic Defect work ICBF is doing where we are collecting DNA, descriptions, and pictures of animals with potential genetic defects from farmers and vets. Once we have enough evidence that a condition has an underlying genetic basis, we try to find the SNP or SNPs responsible for the defect, known as the causative mutation. These potential causative mutations are placed on the next SNP chip we build and get tested each time we genotype an animal. If we have enough cases of a particular disease in a year, then those animals get tested first to see if we really have found the causative mutation. This year, one of the diseases we’ve targeted atresia, or waterbelly. This project is in conjunction with Dr. John Mee and Dr. Orla Keane of Teagasc whose expertise has been a big help on this project.
Please help contribute to these projects by giving ICBF access to your health records through farm software or by recording them online. Also, as we head into Autumn calving, please contact us if you have any calves born with defects. Here is a link to our genetic defect survey (Genetic Defect Survey).

If you’re like more information about what defects are already tested on the IDB chip, click here, and for more information on DNA and genetics, click here.
Any questions about either program can be sent to [email protected]
